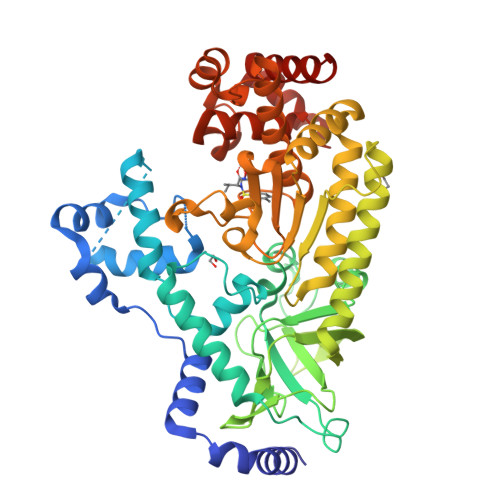
Ligand-induced expansion of the S1' site in the anthrax toxin lethal factor.
Maize, K.M., Kurbanov, E.K., Johnson, R.L., Amin, E.A., Finzel, B.C.(2015) FEBS Lett 589: 3836-3841
- PubMed: 26578066
- DOI: https://doi.org/10.1016/j.febslet.2015.11.005
- Primary Citation of Related Structures:
4XM6, 4XM7, 4XM8 - PubMed Abstract:
The Bacillus anthracis lethal factor (LF) is one component of a tripartite exotoxin partly responsible for persistent anthrax cytotoxicity after initial bacterial infection. Inhibitors of the zinc metalloproteinase have been investigated as potential therapeutic agents, but LF is a challenging target because inhibitors lack sufficient selectivity or possess poor pharmaceutical properties. These structural studies reveal an alternate conformation of the enzyme, induced upon binding of specific inhibitors, that opens a previously unobserved deep pocket termed S1'(∗) which might afford new opportunities to design selective inhibitors that target this subsite.
Organizational Affiliation:
Department of Medicinal Chemistry, University of Minnesota, 308 Harvard St SE, 8-101 Weaver-Densford Hall, Minneapolis, MN 55455, United States.